1. Overview
1.1 Welcome to  PlanT2T
PlanT2T 
Introduction - PlanT2T (The Plant Telomere-to-Telomere Genome Webserver) is an open access platform focusing on plant high-quality assemblies. It aims to provide a centralized and easily accessible platform for researchers to find, analyze and use plant T2T assembly genomic data.
Background - Genome sequencing is crucial for understanding biological traits and evolutionary history [1]. Recently, advanced long-read sequencing has enabled gapless, near-complete or complete chromosome assemblies [2], known as telomere-to-telomere (T2T) assembly (see T2T animation). In plants, T2T assembly has been completed for many species. However, four years after the release of the first plant T2T genome [3], their adoption remains limited. In order to enhance the analysis and management of T2T genomes in plants, we developed PlanT2T.
Websites - We have two websites for PlanT2T ( |
). All data and analysis are provided through the PlanT2T API. Help information for PlanT2T is available at the Help page.
Pipeline - PlanT2T data consists of two parts: user-uploaded and self-collected. All genomes are subjected to quality assessment before basic and deep analysis. Finally, all analyzed data will be permanently stored in PlanT2T.
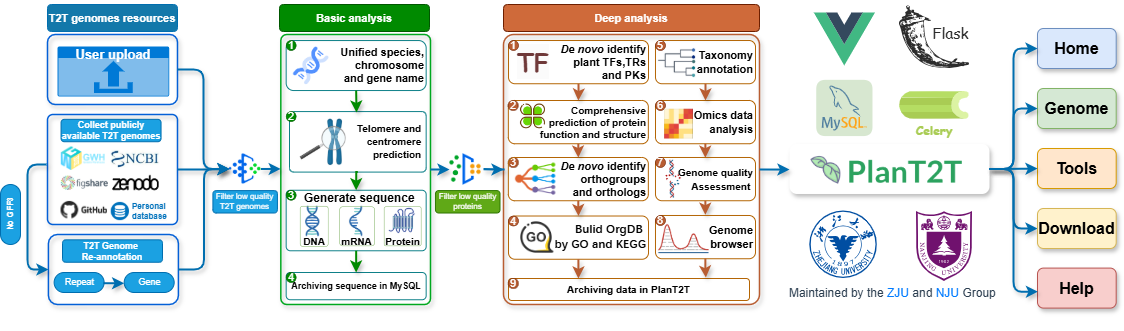
1.2 Main modules
Analysis module - User can upload assembled T2T genomes along with annotation files (GFF3) for analysis. This module provides a series of tools for genome analysis. Finanlly, all results will be displayed with interactive genome pages. User can browse, search, download and share the results at any time.
Sarch module - PlanT2T collected and curated over 180 T2T genomes, including newly annotated ones. In addition to employing the same analytical method mentioned above, we also analyzed the relationships within each gene orthogroup (OG), as well as a wealth of omics data.
1.3 Comparison
| High quality 1 |
Automated analysis 2 |
Interactive visualization 3 |
Query search engine 4 |
Fully open 5 |
|
|---|---|---|---|---|---|
| PlanT2T | ✔ | ✔ | ✔ | ✔ | ✔ |
| Phytozome | ⚪ | ❌ | ✔ | ✔ | ⚪ |
| Ensembl Plants | ⚪ | ❌ | ✔ | ✔ | ✔ |
| NCBI Genome | ⚪ | ❌ | ✔ | ✔ | ✔ |
| GWH | ⚪ | ❌ | ❌ | ❌ | ⚪ |
| PubPlant | ⚪ | ❌ | ❌ | ❌ | ✔ |
| Plant GARDEN | ⚪ | ❌ | ✔ | ✔ | ⚪ |
| PlantGIR | ⚪ | ❌ | ❌ | ❌ | ✔ |
| IMP | ⚪ | ❌ | ✔ | ✔ | ✔ |
✔: Yes, ❌: No, ⚪: Partial
1 High quality data - PlanT2T only provides telomere-to-telomere, gapfree, gapless, near-complete chromosome assemblies for plant species.
2 Automated analysis - PlanT2T automatically analyzes the uploaded T2T genome data (No login and review required) and displays the analysis results interactively to help users better browse, analyze, and share plant T2T assembly data.
3 Interactive visualization - PlanT2T provides interactive visualization of each species genome, gene annotation, gene families, omics data and other information.
4 Query search engine - PlanT2T provides a powerful search engine for users to search for specific genes, gene functions, gene families, and other information.
5 Fully open - PlanT2T is an open access platform that provides all data and analysis results for free.
1.4 More questions?
If you have any questions or suggestions, please feel free to Contact us or submit an issue on GitHub.