2.1 Usage
2.1.1 Introduction
Here, we provides a detailed explanation of the Genome Analysis module available on the PlantT2T platform. The module allows users to upload T2T genome sequences and annotation files(GFF3)for comprehensive analysis. 🐱🏍Go to the page

Input Requirements
To use the Genome Analysis module, the following inputs are required:
Genome File - Upload the T2T genome in FASTA format (accepts .fa, .fasta, .fna, .gz, .tar.gz, .zip).
Annotation File - Provide a GFF3 file containing gene annotations (accepts .gff3, .gff, .gz, .tar.gz, .zip).
Latin Name - Specify the Latin name of the species (e.g., Oryza sativa. It is preferable that the information can be accurately located on NCBI’s Taxonomy Database)
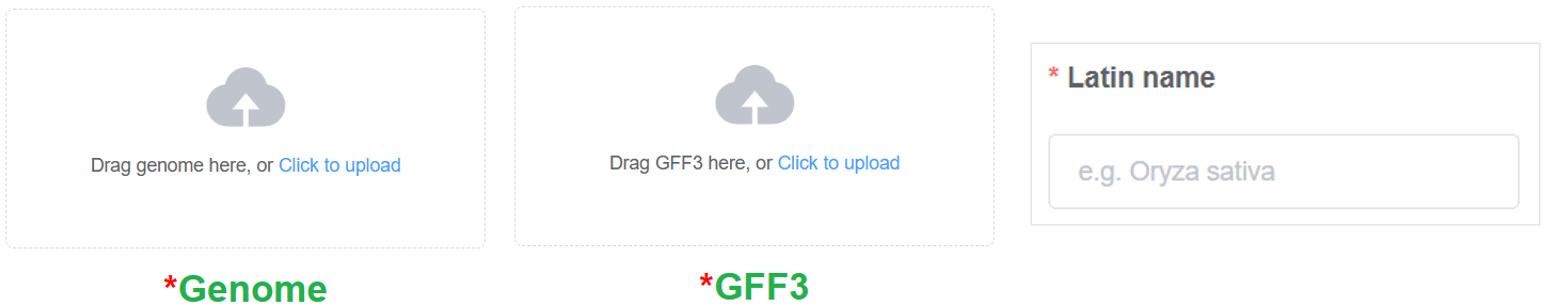
Orther inputs are optional. However, provide more information can enrich your genome page.
2.1.2 Getting Started
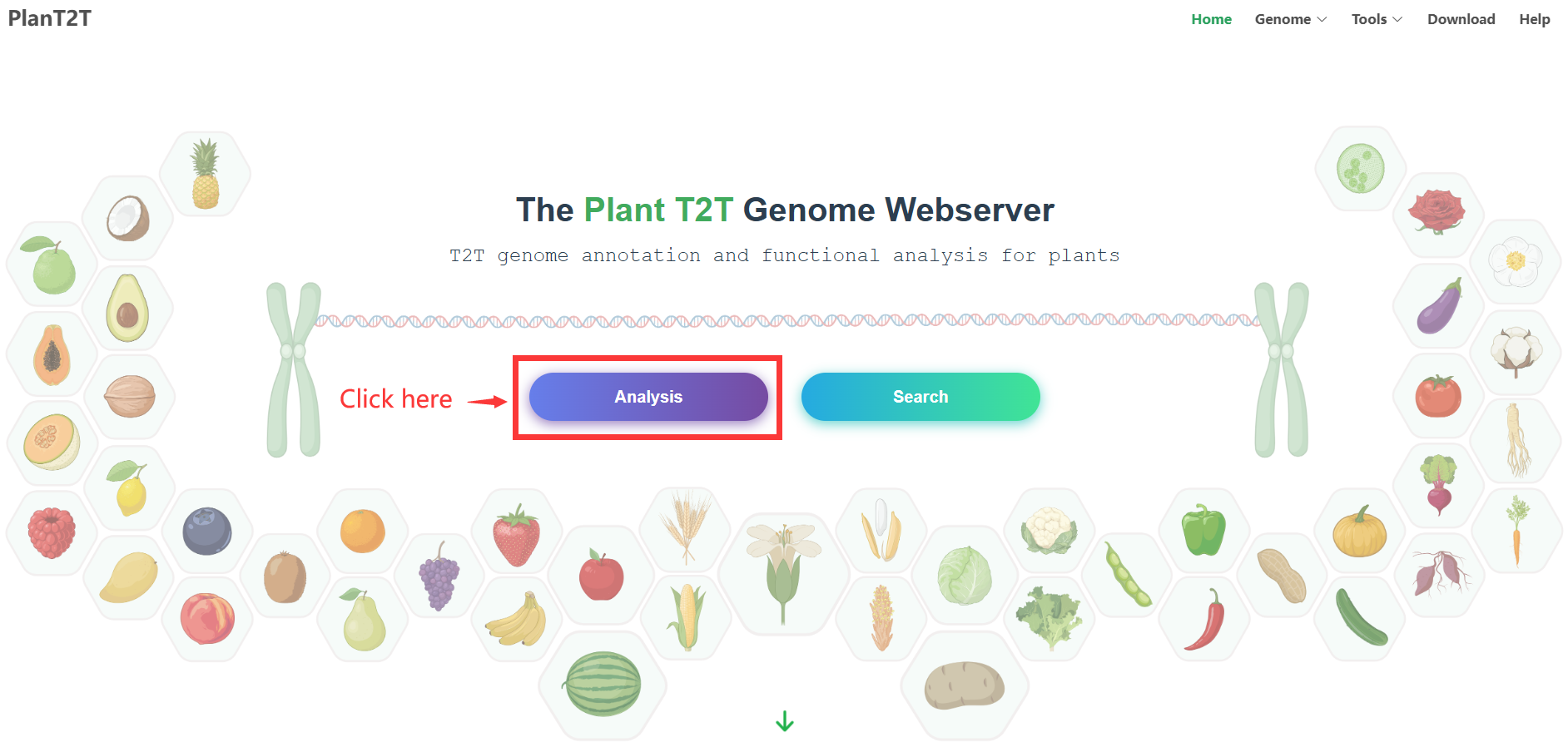
1. Enter the Analysis Module
Navigate to the PlantT2T homepage and select the Analysis module.
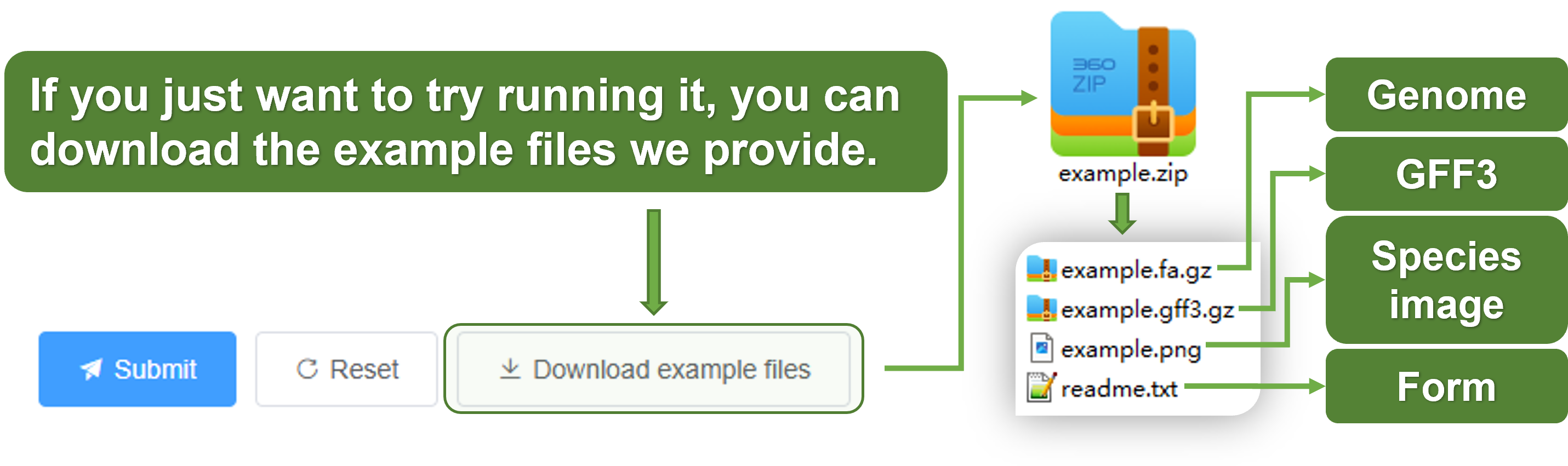
Download the example files (optional)
Since analyzing an entire genome takes a long time, here we provide example files to help you quickly experience this module. You can download the example files here or by clicking the Download Example Files button on the Analysis page.
- genome file (example.fa.gz),
- GFF3 annotation file (example.gff3.gz),
- species image (example.png),
- form example file (readme.txt).
These can help you get started quickly. The runtime is approximately 30 minutes. We have completed the example genome analysis. You can view the results by clicking the here.
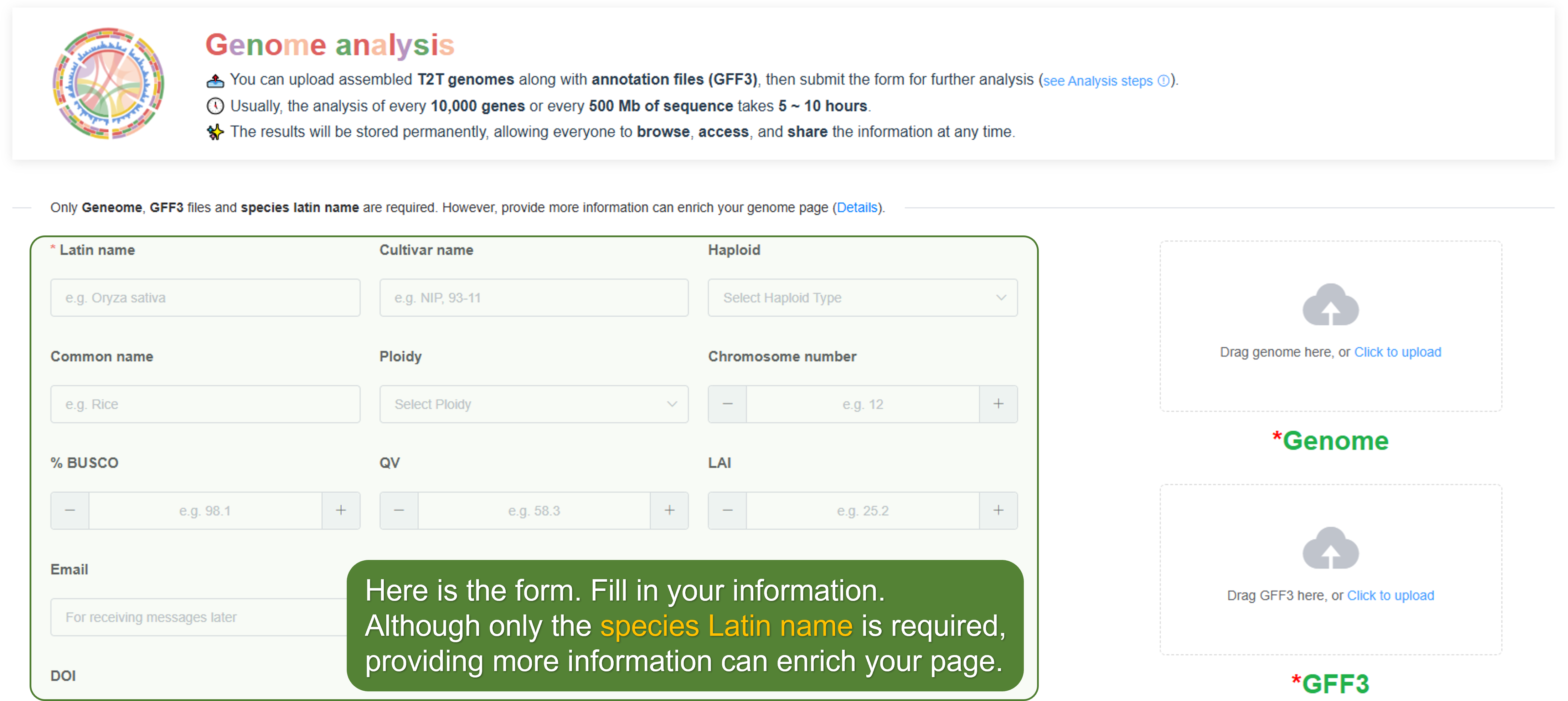
2. Fill in the form
Provide the necessary information:
Latin Name - Enter the formal scientific name of the species.
Optional fields - Other fields are optional, but providing more information can enrich your genome page.
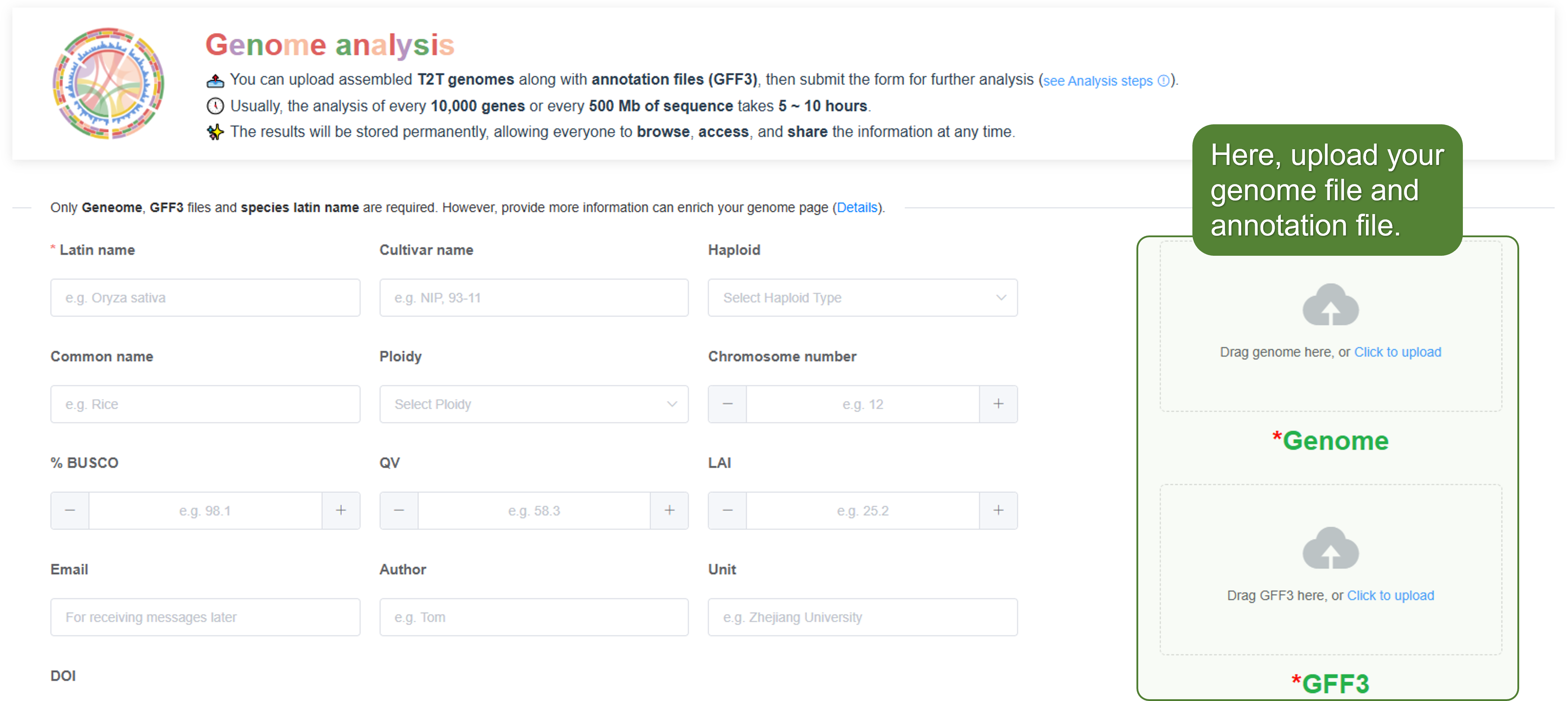
3. Upload files
Drop or click to upload the required files:
- Genome sequence file (FASTA format).
- Annotation file (GFF3 format).
- Species image (PNG format) - optional.
Before uploading, you can check the genome and GFF3 files to ensure they are in the correct format and are high-quality T2T assemblies. Otherwise, if your files do not pass our checks, the analysis will terminate, and you will need to make repeated modifications, which can take a lot of time. Therefore, you can use our script to perform the checks in advance.
See code for checking genome and GFF3 files
# Download the genomeCheck script
wget https://biobigdata.nju.edu.cn/plant2t/script/genomeCheck
chmod +x genomeCheck
# Run the script
genome=example.fa.gz
annotation=example.gff3.gz
./genomeCheck -g ${genome} -a ${annotation}After running the script smoothly, you will see the following output:
Pass GFF3 check: GFF3 annotations match the FASTA sequences.
Pass protein check: the rate of low-quality protein sequences is 1.34228%.
Pass gaps check: total 1 gaps were found in your genome.
Pass sequences filter: all sequences are longer than 1000000 bp.
Pass Chromosome names chack: Chromosome names start with 'Chr', no need to rename.
Two files were generated:
/mnt/c/Users/haoyu/Desktop/example.fa.new.fa
/mnt/c/Users/haoyu/Desktop/example.gff3.new.gff3
You can upload them to PlanT2T for further analysis.
Pass all check!If the output shows some errors, please fix them and rerun the script. If you cannot fix them, please open an issue on the GitHub.
4. Submit
After filling all fields, click the Submit button.
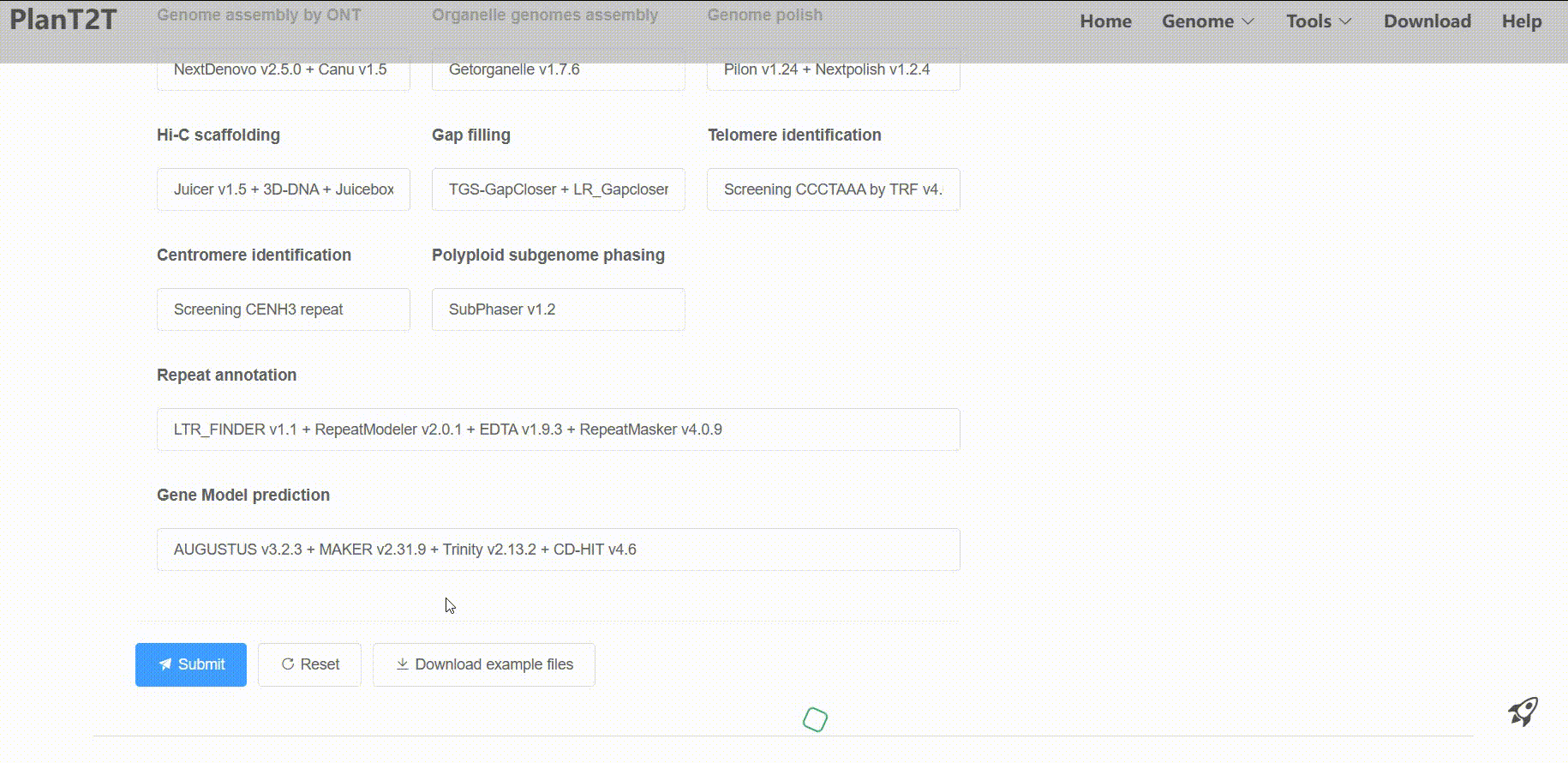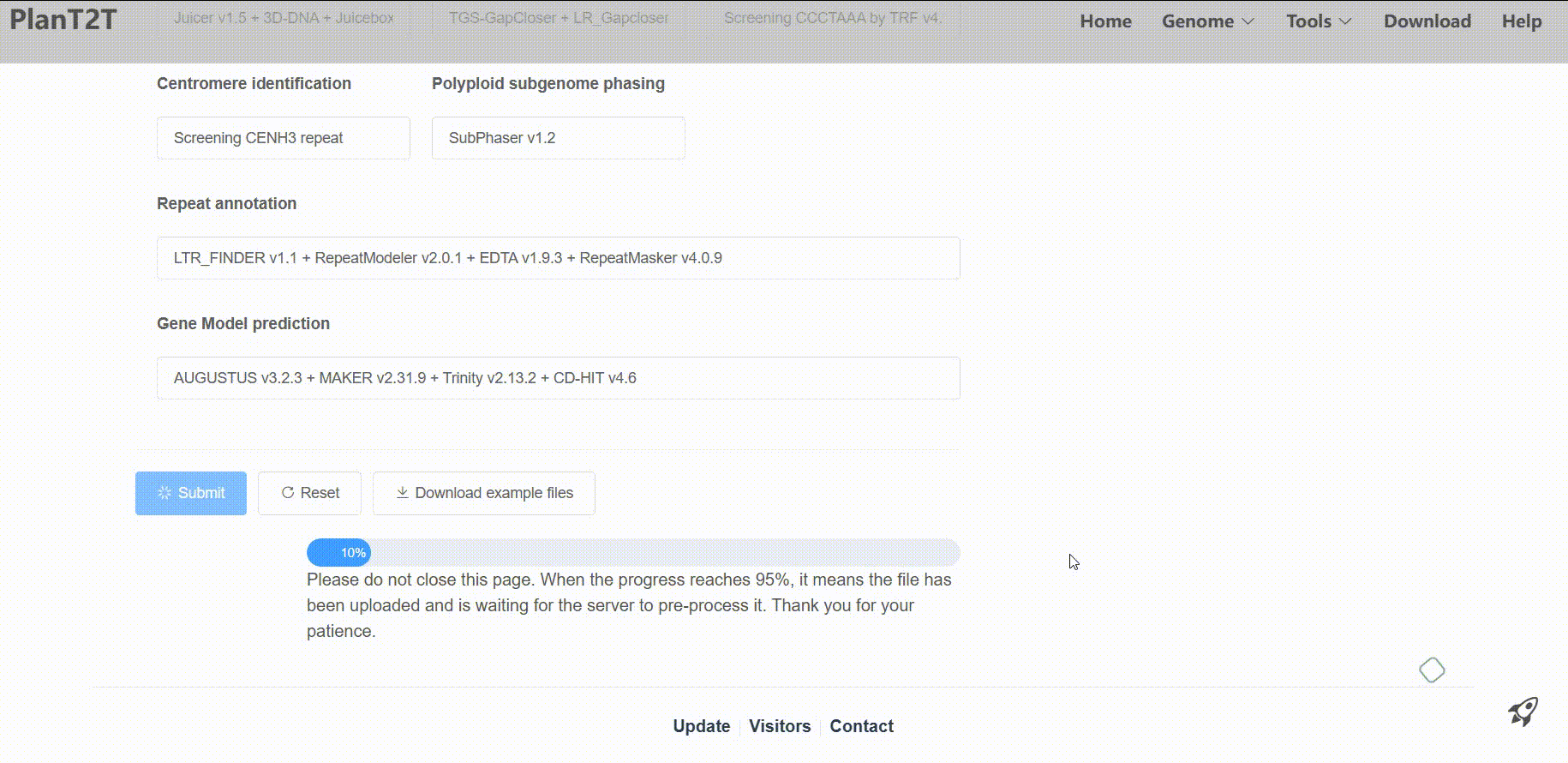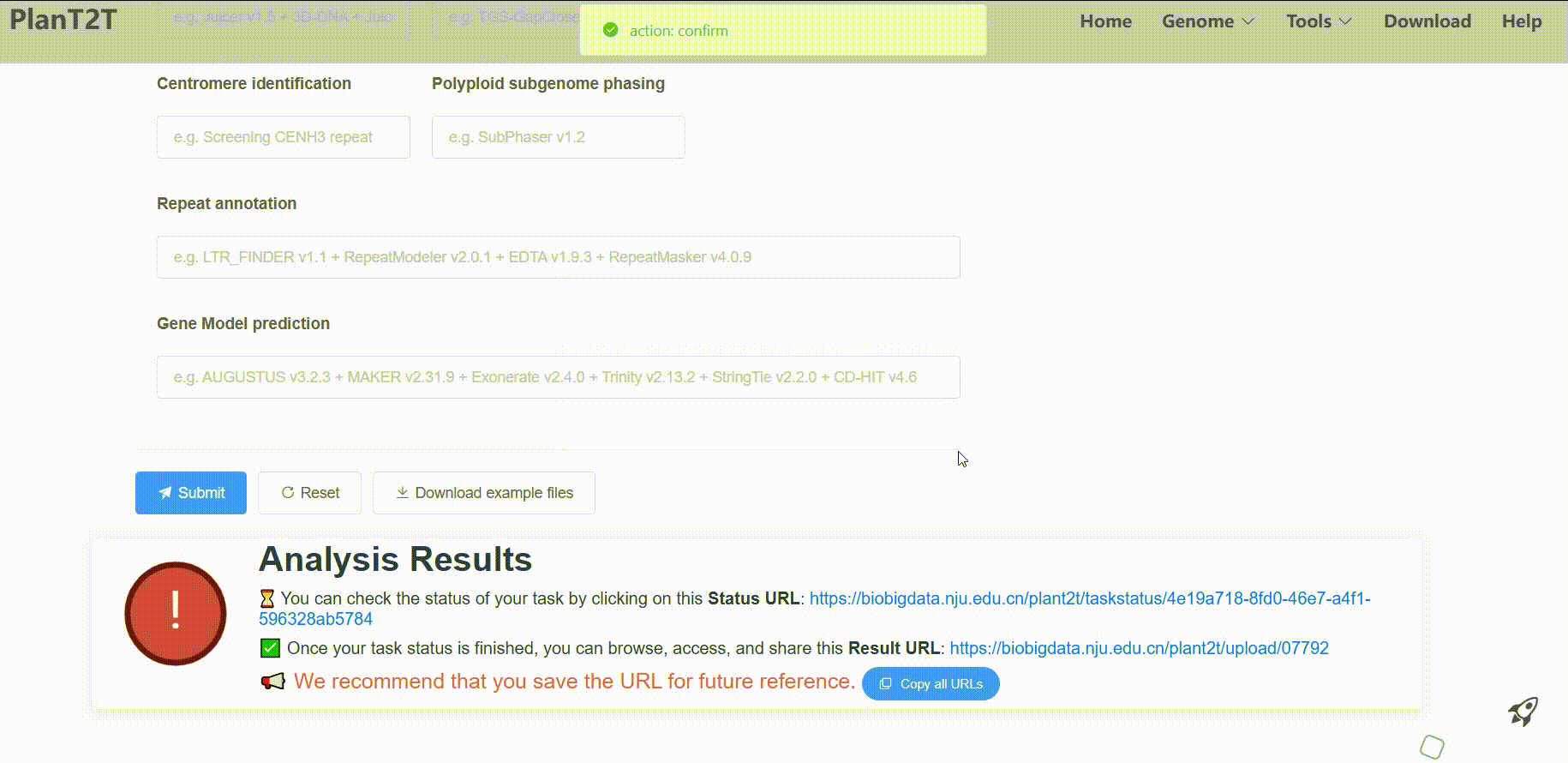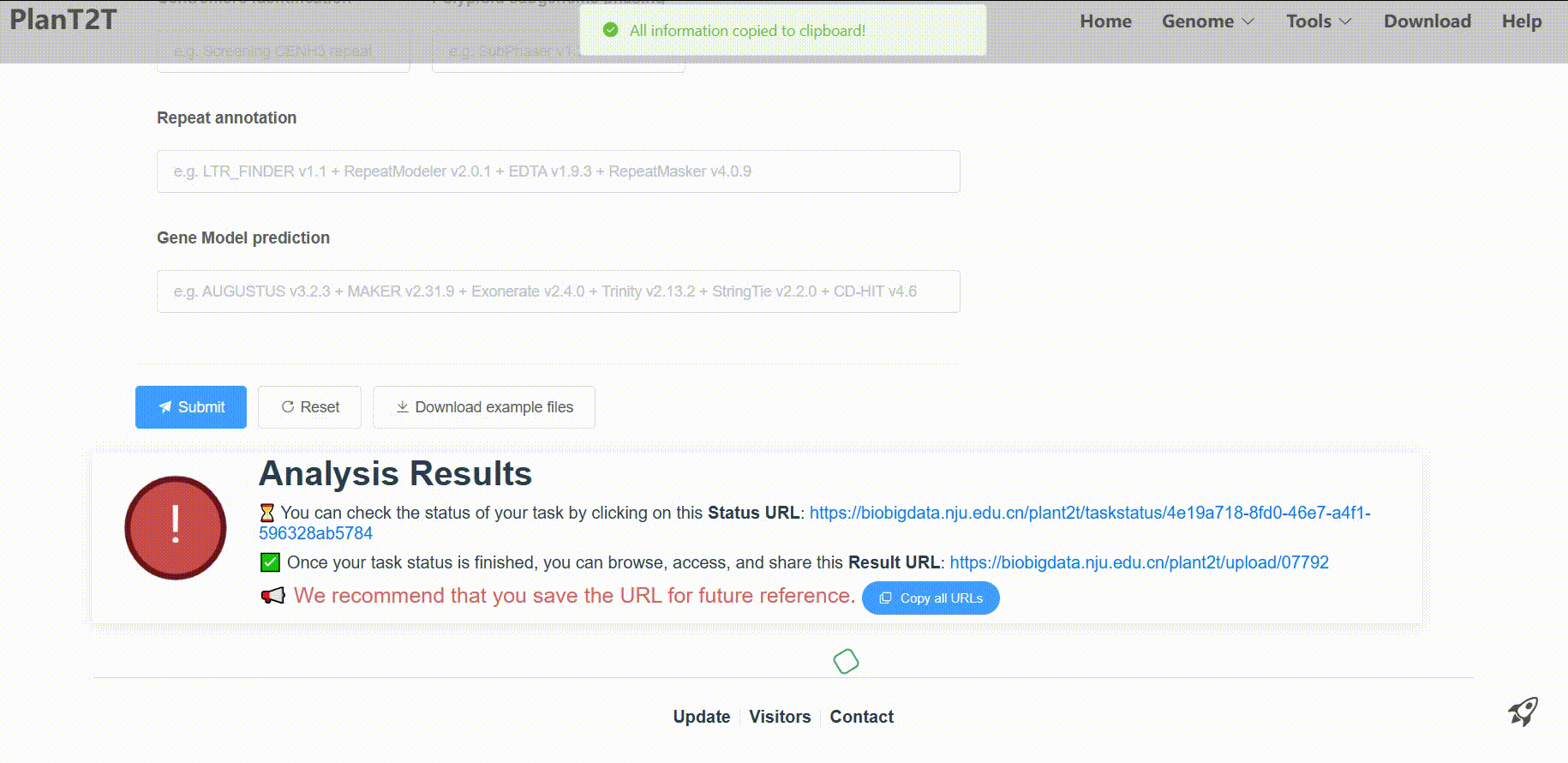
Then, the platform will process your data for analysis. Processing times vary based on genome size, typically taking 5–10 hours for 10,000 genes or 500 Mb sequences. In order to check the status of your task in real-time, please save the Status URL and Result URL after submission.
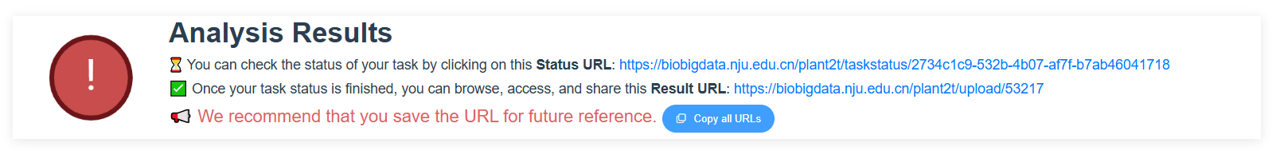
We have completed the example genome analysis. You can view the results by clicking here.
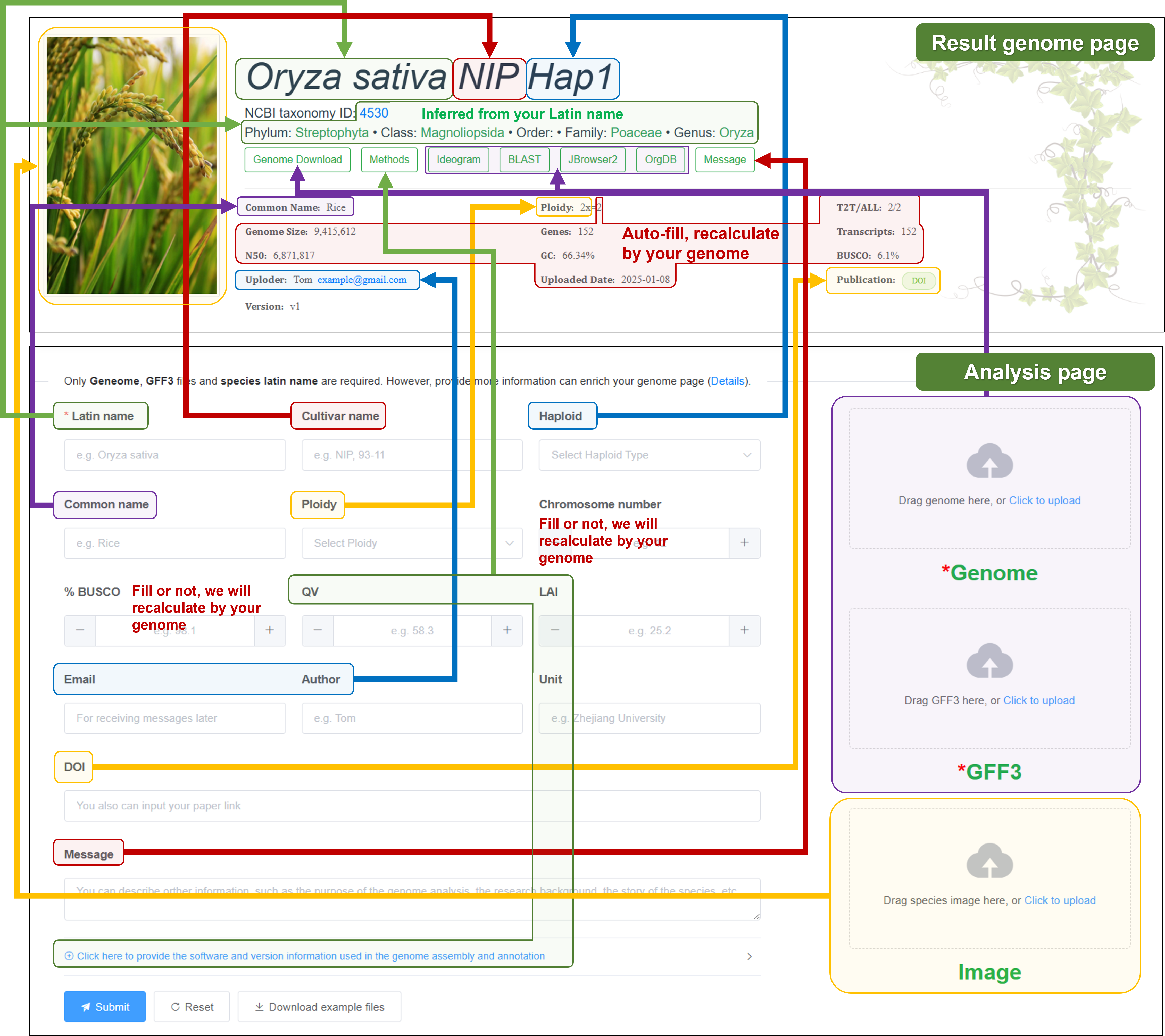
5. Mapping your form to the genome page
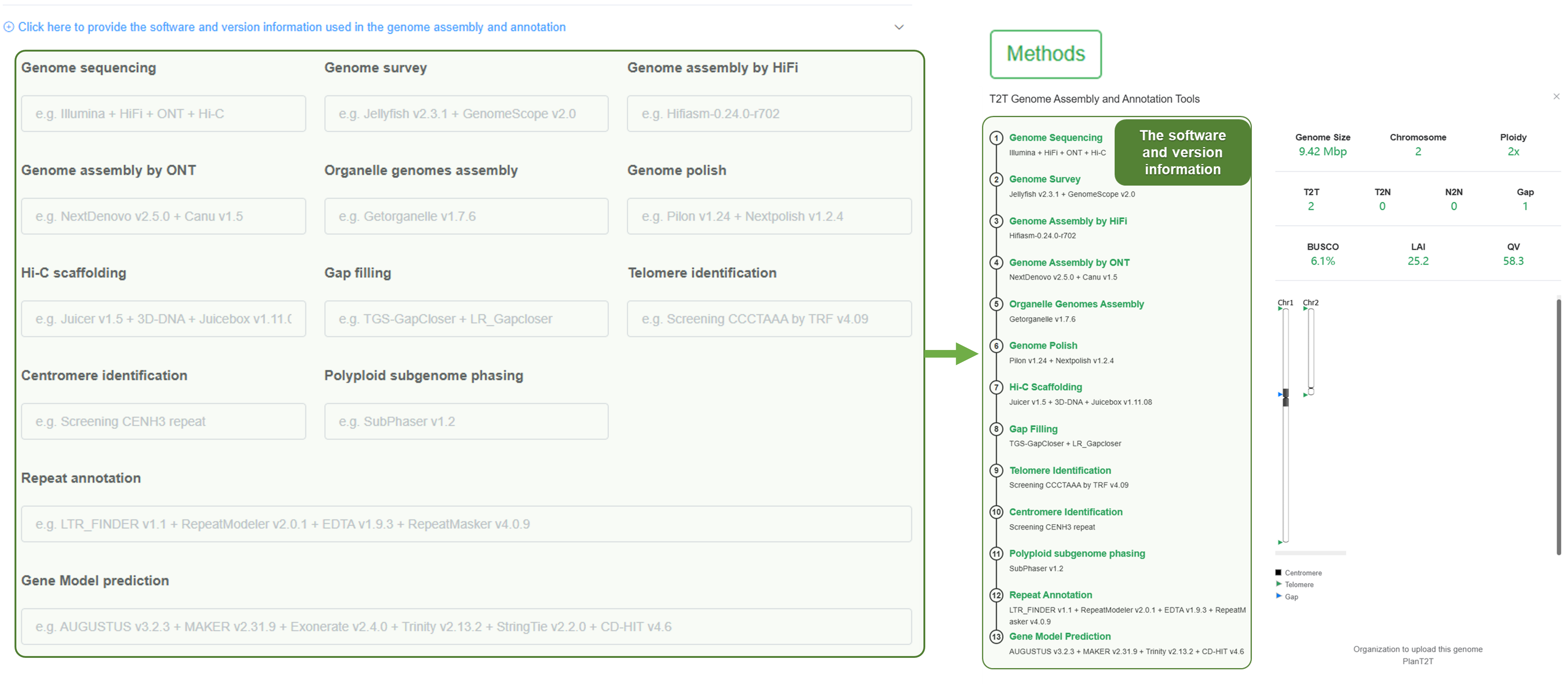
Here, we will show you the corresponding part of the genome page for each item you fill in, helping you judge whether the content meets your expectations.
2.1.3 Troubleshooting and Support
File Upload - Ensure 1. genome and annotation files are in the correct format. 2. genome are high-quality T2T assemblies. 3. files are not corrupted.
Analysis Delays - Large genome sizes (>1Gb) or A large number of protein sequences (>10k) may take longer to process.
Incomplete Results - If the analysis is incomplete or interrupted please check the message for errors. Then modify the input files and re-upload if needed.
Contact support - For further assistance, please contact us on GitHub or Email .