4.4 KEGG Enrichment
4.4.1 Usage
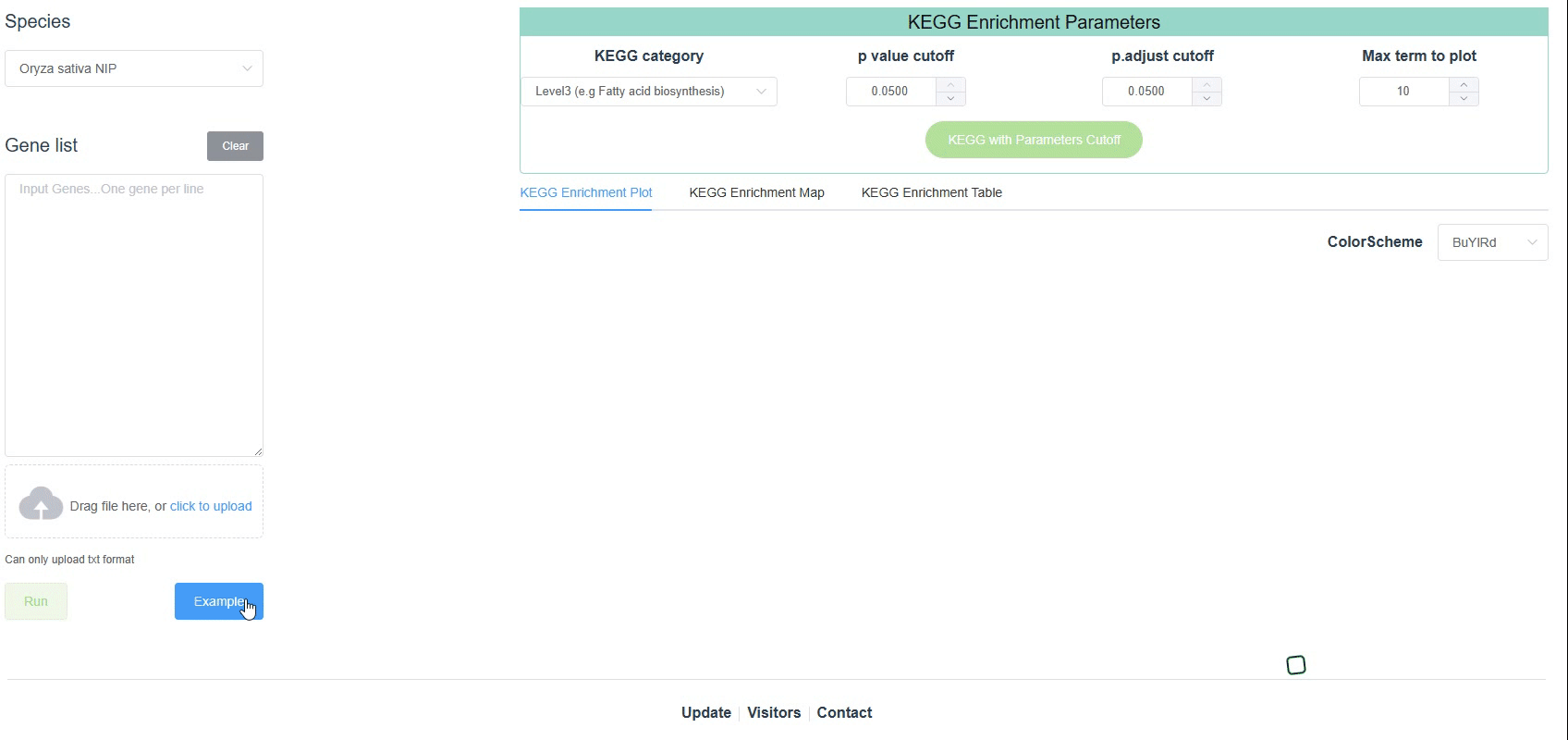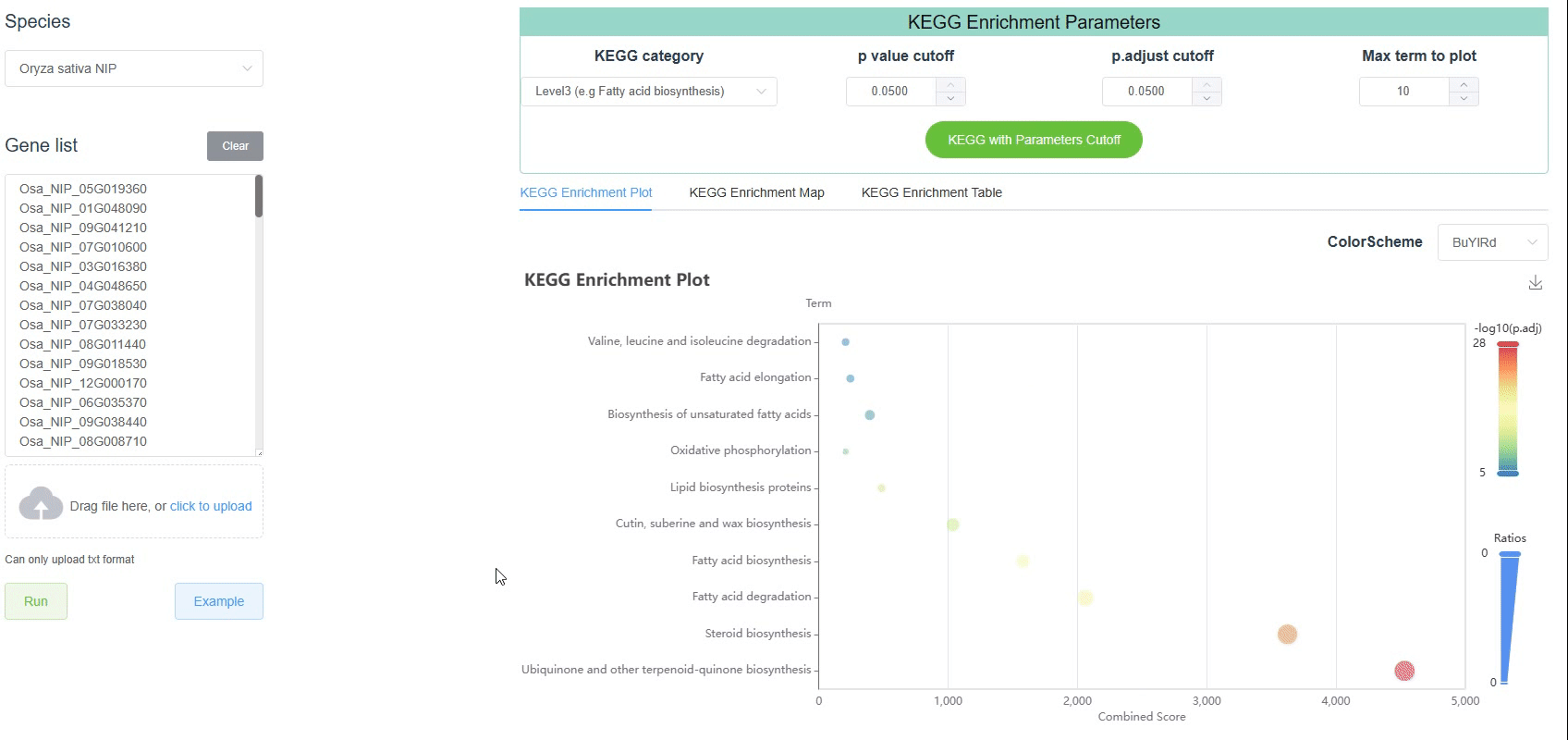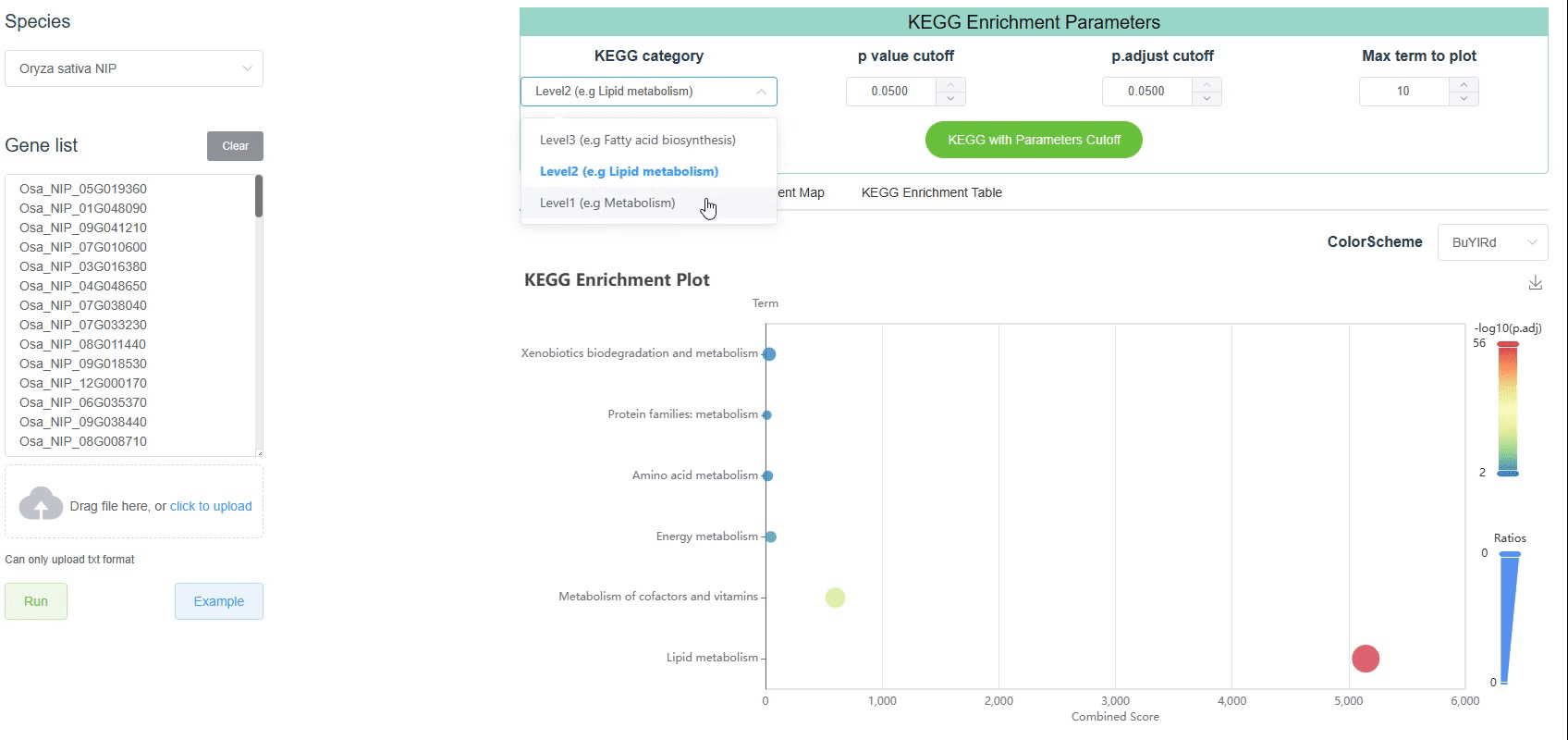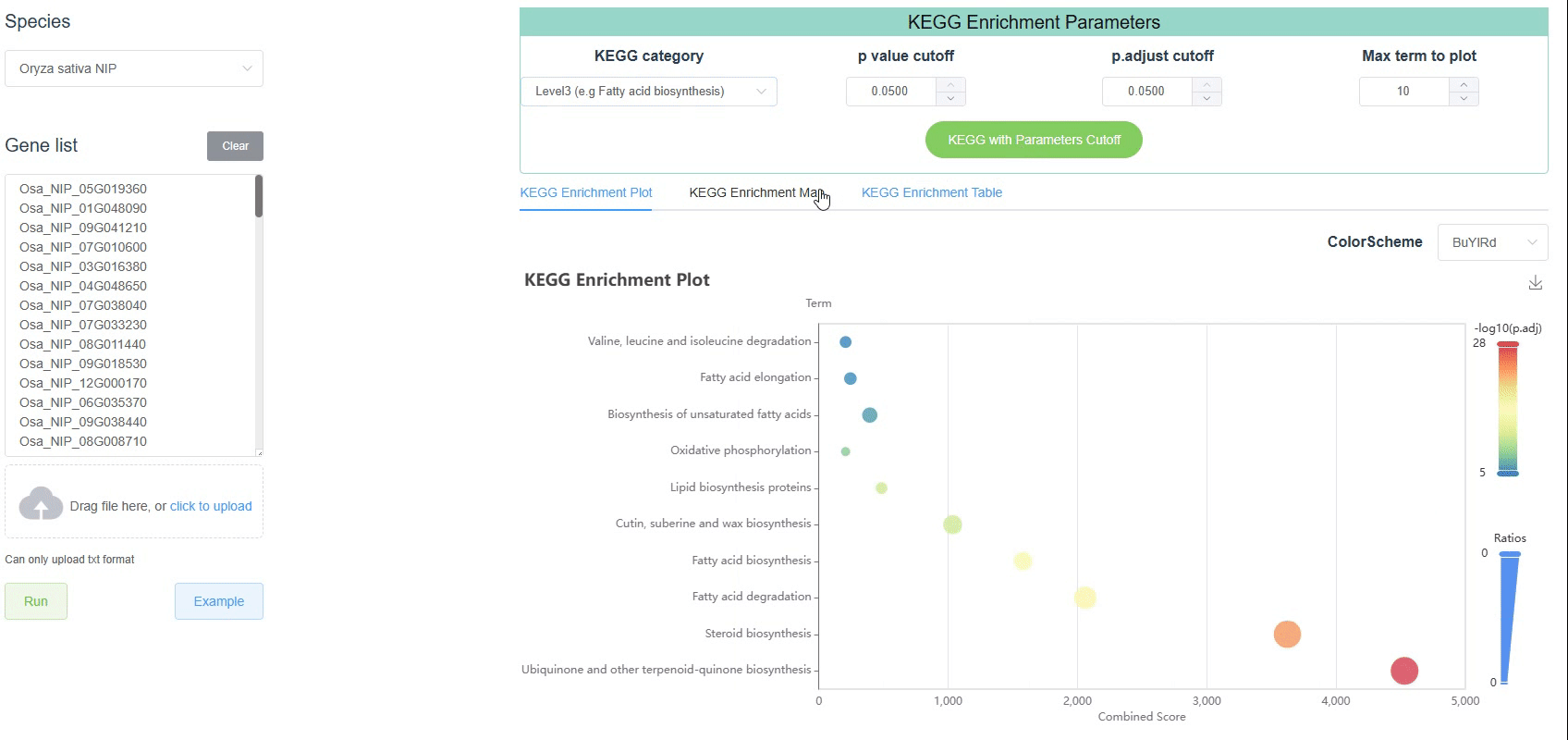
Like the GO enrichment tool, the KEGG enrichment tool in PlanT2T helps users identify biological pathways that are statistically overrepresented among a list of differentially expressed genes. Users can upload the PlanT2T ID in the correct format and the program will perform the KEGG enrichment.
4.4.2 Level description
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway consists of three levels. Inorder to better understand the KEGG enrichment results, we select plant related pathways for display. The three levels are as follows:
Level 1: The first level is the top-level pathway, which is the most general pathway classification. Here, only 5 pathways are displayed.
Metabolism
Genetic Information Processing
Environmental Information Processing
Cellular Processes
Brite HierarchiesLevel 2: The second level is the middle-level pathway, which is a more detailed classification of the pathway. Here, only 25 pathways are displayed.
Carbohydrate metabolism
Energy metabolism
Lipid metabolism
Nucleotide metabolism
Amino acid metabolism
Metabolism of other amino acids
Glycan biosynthesis and metabolism
Metabolism of cofactors and vitamins
Metabolism of terpenoids and polyketides
Biosynthesis of other secondary metabolites
Xenobiotics biodegradation and metabolism
Transcription
Translation
Folding, sorting and degradation
Replication and repair
Chromosome
Membrane transport
Signal transduction
Transport and catabolism
Cell growth and death
Cell motility
Protein families: metabolism
Protein families: genetic information processing
Protein families: signaling and cellular processes
RNA familyLevel 3: The third level is the lowest level pathway, which is the most detailed classification of the pathway. Here, 253 pathways are displayed.
ko00010 Glycolysis / Gluconeogenesis
ko00020 Citrate cycle (TCA cycle)
ko00030 Pentose phosphate pathway
ko00040 Pentose and glucuronate interconversions
ko00051 Fructose and mannose metabolism
ko00052 Galactose metabolism
ko00053 Ascorbate and aldarate metabolism
ko00500 Starch and sucrose metabolism
ko00520 Amino sugar and nucleotide sugar metabolism
ko00620 Pyruvate metabolism
ko00630 Glyoxylate and dicarboxylate metabolism
ko00640 Propanoate metabolism
ko00650 Butanoate metabolism
ko00660 C5-Branched dibasic acid metabolism
ko00562 Inositol phosphate metabolism
ko00190 Oxidative phosphorylation
ko00195 Photosynthesis
ko00196 Photosynthesis - antenna proteins
ko00710 Carbon fixation by Calvin cycle
ko00720 Other carbon fixation pathways
ko00680 Methane metabolism
ko00910 Nitrogen metabolism
ko00920 Sulfur metabolism
ko00061 Fatty acid biosynthesis
ko00062 Fatty acid elongation
ko00071 Fatty acid degradation
ko00073 Cutin, suberine and wax biosynthesis
ko00100 Steroid biosynthesis
ko00120 Primary bile acid biosynthesis
ko00121 Secondary bile acid biosynthesis
ko00561 Glycerolipid metabolism
ko00564 Glycerophospholipid metabolism
ko00565 Ether lipid metabolism
ko00600 Sphingolipid metabolism
ko00591 Linoleic acid metabolism
ko00592 alpha-Linolenic acid metabolism
ko01040 Biosynthesis of unsaturated fatty acids
ko00230 Purine metabolism
ko00240 Pyrimidine metabolism
ko00250 Alanine, aspartate and glutamate metabolism
ko00260 Glycine, serine and threonine metabolism
ko00270 Cysteine and methionine metabolism
ko00280 Valine, leucine and isoleucine degradation
ko00290 Valine, leucine and isoleucine biosynthesis
ko00300 Lysine biosynthesis
ko00310 Lysine degradation
ko00220 Arginine biosynthesis
ko00330 Arginine and proline metabolism
ko00340 Histidine metabolism
ko00350 Tyrosine metabolism
ko00360 Phenylalanine metabolism
ko00380 Tryptophan metabolism
ko00400 Phenylalanine, tyrosine and tryptophan biosynthesis
ko00410 beta-Alanine metabolism
ko00430 Taurine and hypotaurine metabolism
ko00440 Phosphonate and phosphinate metabolism
ko00450 Selenocompound metabolism
ko00460 Cyanoamino acid metabolism
ko00470 D-Amino acid metabolism
ko00480 Glutathione metabolism
ko00510 N-Glycan biosynthesis
ko00513 Various types of N-glycan biosynthesis
ko00512 Mucin type O-glycan biosynthesis
ko00515 Mannose type O-glycan biosynthesis
ko00514 Other types of O-glycan biosynthesis
ko00532 Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate
ko00534 Glycosaminoglycan biosynthesis - heparan sulfate / heparin
ko00533 Glycosaminoglycan biosynthesis - keratan sulfate
ko00531 Glycosaminoglycan degradation
ko00563 Glycosylphosphatidylinositol (GPI)-anchor biosynthesis
ko00601 Glycosphingolipid biosynthesis - lacto and neolacto series
ko00603 Glycosphingolipid biosynthesis - globo and isoglobo series
ko00604 Glycosphingolipid biosynthesis - ganglio series
ko00511 Other glycan degradation
ko00540 Lipopolysaccharide biosynthesis
ko00542 O-Antigen repeat unit biosynthesis
ko00541 O-Antigen nucleotide sugar biosynthesis
ko00550 Peptidoglycan biosynthesis
ko00552 Teichoic acid biosynthesis
ko00571 Lipoarabinomannan (LAM) biosynthesis
ko00572 Arabinogalactan biosynthesis - Mycobacterium
ko00543 Exopolysaccharide biosynthesis
ko00730 Thiamine metabolism
ko00740 Riboflavin metabolism
ko00750 Vitamin B6 metabolism
ko00760 Nicotinate and nicotinamide metabolism
ko00770 Pantothenate and CoA biosynthesis
ko00780 Biotin metabolism
ko00785 Lipoic acid metabolism
ko00790 Folate biosynthesis
ko00670 One carbon pool by folate
ko00830 Retinol metabolism
ko00860 Porphyrin metabolism
ko00130 Ubiquinone and other terpenoid-quinone biosynthesis
ko00900 Terpenoid backbone biosynthesis
ko00902 Monoterpenoid biosynthesis
ko00909 Sesquiterpenoid and triterpenoid biosynthesis
ko00904 Diterpenoid biosynthesis
ko00906 Carotenoid biosynthesis
ko00905 Brassinosteroid biosynthesis
ko00908 Zeatin biosynthesis
ko00903 Limonene degradation
ko00907 Pinene, camphor and geraniol degradation
ko01052 Type I polyketide structures
ko00522 Biosynthesis of 12-, 14- and 16-membered macrolides
ko01051 Biosynthesis of ansamycins
ko01059 Biosynthesis of enediyne antibiotics
ko01056 Biosynthesis of type II polyketide backbone
ko01057 Biosynthesis of type II polyketide products
ko00253 Tetracycline biosynthesis
ko00523 Polyketide sugar unit biosynthesis
ko01054 Nonribosomal peptide structures
ko01053 Biosynthesis of siderophore group nonribosomal peptides
ko01055 Biosynthesis of vancomycin group antibiotics
ko00940 Phenylpropanoid biosynthesis
ko00945 Stilbenoid, diarylheptanoid and gingerol biosynthesis
ko00941 Flavonoid biosynthesis
ko00944 Flavone and flavonol biosynthesis
ko00942 Anthocyanin biosynthesis
ko00943 Isoflavonoid biosynthesis
ko00946 Degradation of flavonoids
ko00901 Indole alkaloid biosynthesis
ko00403 Indole diterpene alkaloid biosynthesis
ko00950 Isoquinoline alkaloid biosynthesis
ko00960 Tropane, piperidine and pyridine alkaloid biosynthesis
ko00996 Biosynthesis of various alkaloids
ko00232 Caffeine metabolism
ko00965 Betalain biosynthesis
ko00966 Glucosinolate biosynthesis
ko00402 Benzoxazinoid biosynthesis
ko00311 Penicillin and cephalosporin biosynthesis
ko00332 Carbapenem biosynthesis
ko00261 Monobactam biosynthesis
ko00331 Clavulanic acid biosynthesis
ko00521 Streptomycin biosynthesis
ko00524 Neomycin, kanamycin and gentamicin biosynthesis
ko00525 Acarbose and validamycin biosynthesis
ko00401 Novobiocin biosynthesis
ko00404 Staurosporine biosynthesis
ko00405 Phenazine biosynthesis
ko00333 Prodigiosin biosynthesis
ko00254 Aflatoxin biosynthesis
ko00998 Biosynthesis of various antibiotics
ko00999 Biosynthesis of various plant secondary metabolites
ko00997 Biosynthesis of various other secondary metabolites
ko00362 Benzoate degradation
ko00627 Aminobenzoate degradation
ko00364 Fluorobenzoate degradation
ko00625 Chloroalkane and chloroalkene degradation
ko00361 Chlorocyclohexane and chlorobenzene degradation
ko00623 Toluene degradation
ko00622 Xylene degradation
ko00633 Nitrotoluene degradation
ko00642 Ethylbenzene degradation
ko00643 Styrene degradation
ko00791 Atrazine degradation
ko00930 Caprolactam degradation
ko00363 Bisphenol degradation
ko00621 Dioxin degradation
ko00626 Naphthalene degradation
ko00624 Polycyclic aromatic hydrocarbon degradation
ko00365 Furfural degradation
ko00984 Steroid degradation
ko00980 Metabolism of xenobiotics by cytochrome P450
ko00982 Drug metabolism - cytochrome P450
ko03020 RNA polymerase
ko03022 Basal transcription factors
ko03040 Spliceosome
ko03010 Ribosome
ko00970 Aminoacyl-tRNA biosynthesis
ko03013 Nucleocytoplasmic transport
ko03015 mRNA surveillance pathway
ko03008 Ribosome biogenesis in eukaryotes
ko03060 Protein export
ko04141 Protein processing in endoplasmic reticulum
ko04130 SNARE interactions in vesicular transport
ko04120 Ubiquitin mediated proteolysis
ko04122 Sulfur relay system
ko03050 Proteasome
ko03018 RNA degradation
ko03030 DNA replication
ko03410 Base excision repair
ko03420 Nucleotide excision repair
ko03430 Mismatch repair
ko03440 Homologous recombination
ko03450 Non-homologous end-joining
ko03082 ATP-dependent chromatin remodeling
ko03083 Polycomb repressive complex
ko02010 ABC transporters
ko02020 Two-component system
ko04010 MAPK signaling pathway
ko04016 MAPK signaling pathway - plant
ko04020 Calcium signaling pathway
ko04070 Phosphatidylinositol signaling system
ko04072 Phospholipase D signaling pathway
ko04071 Sphingolipid signaling pathway
ko04022 cGMP-PKG signaling pathway
ko04151 PI3K-Akt signaling pathway
ko04152 AMPK signaling pathway
ko04150 mTOR signaling pathway
ko04075 Plant hormone signal transduction
ko04144 Endocytosis
ko04142 Lysosome
ko04146 Peroxisome
ko04110 Cell cycle
ko04210 Apoptosis
ko04216 Ferroptosis
ko04218 Cellular senescence
ko04814 Motor proteins
ko01000 Enzymes
ko01001 Protein kinases
ko01009 Protein phosphatases and associated proteins
ko01002 Peptidases and inhibitors
ko01003 Glycosyltransferases
ko01004 Lipid biosynthesis proteins
ko01008 Polyketide biosynthesis proteins
ko01006 Prenyltransferases
ko01007 Amino acid related enzymes
ko00199 Cytochrome P450
ko00194 Photosynthesis proteins
ko03000 Transcription factors
ko03021 Transcription machinery
ko03019 Messenger RNA biogenesis
ko03041 Spliceosome
ko03011 Ribosome
ko03009 Ribosome biogenesis
ko03016 Transfer RNA biogenesis
ko03012 Translation factors
ko03110 Chaperones and folding catalysts
ko04131 Membrane trafficking
ko04121 Ubiquitin system
ko03051 Proteasome
ko03032 DNA replication proteins
ko03036 Chromosome and associated proteins
ko03400 DNA repair and recombination proteins
ko03029 Mitochondrial biogenesis
ko02000 Transporters
ko02044 Secretion system
ko02022 Two-component system
ko04812 Cytoskeleton proteins
ko04147 Exosome
ko04030 G protein-coupled receptors
ko04050 Cytokine receptors
ko04054 Pattern recognition receptors
ko03310 Nuclear receptors
ko04040 Ion channels
ko04031 GTP-binding proteins
ko04515 Cell adhesion molecules
ko00535 Proteoglycans
ko00536 Glycosaminoglycan binding proteins
ko00537 Glycosylphosphatidylinositol (GPI)-anchored proteins
ko04091 Lectins
ko03100 Non-coding RNAs