Overview
As a database dedicated to plant genomes, our goal is to offer a wide range of valuable tools and a wealth of data for our users to utilize. We are pleased to provide a comprehensive user guide for our database, and we hope that you will find it both informative and enjoyable to use.
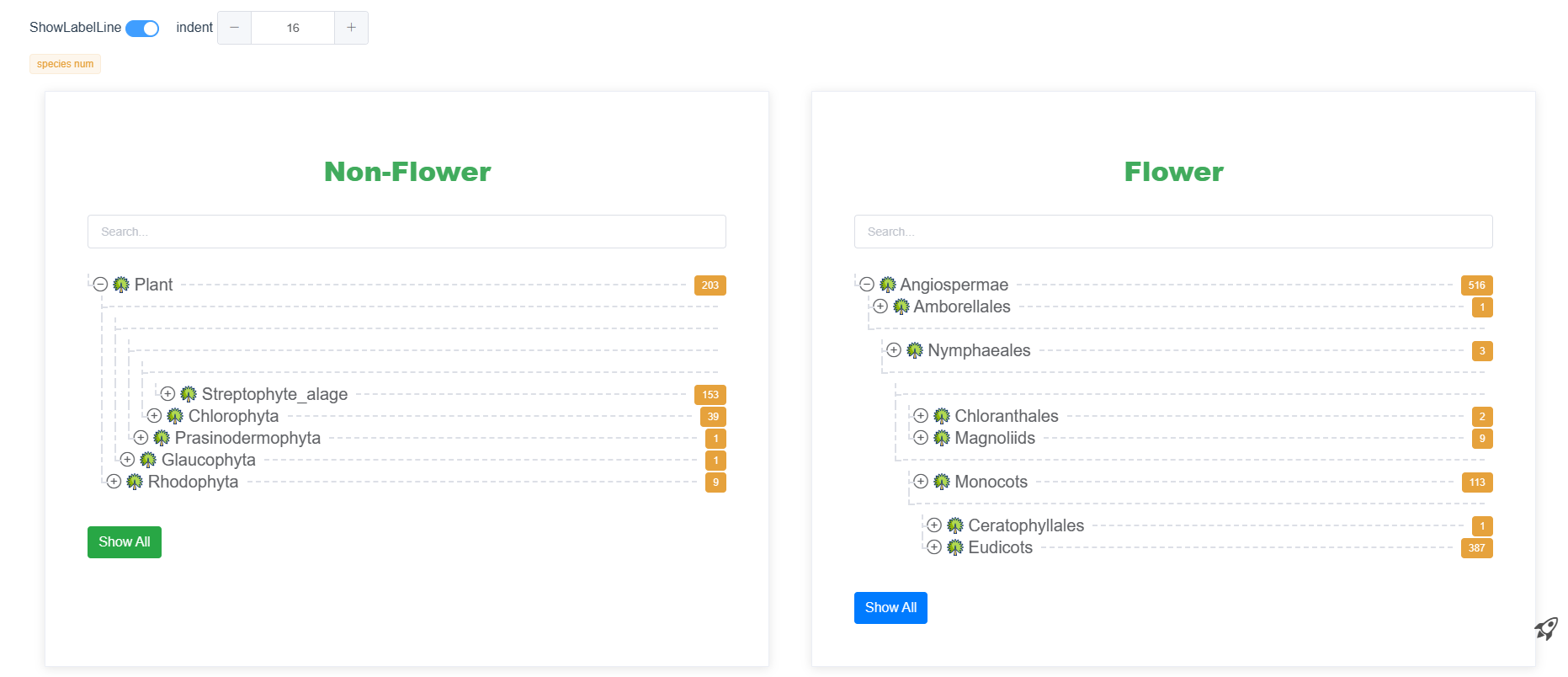
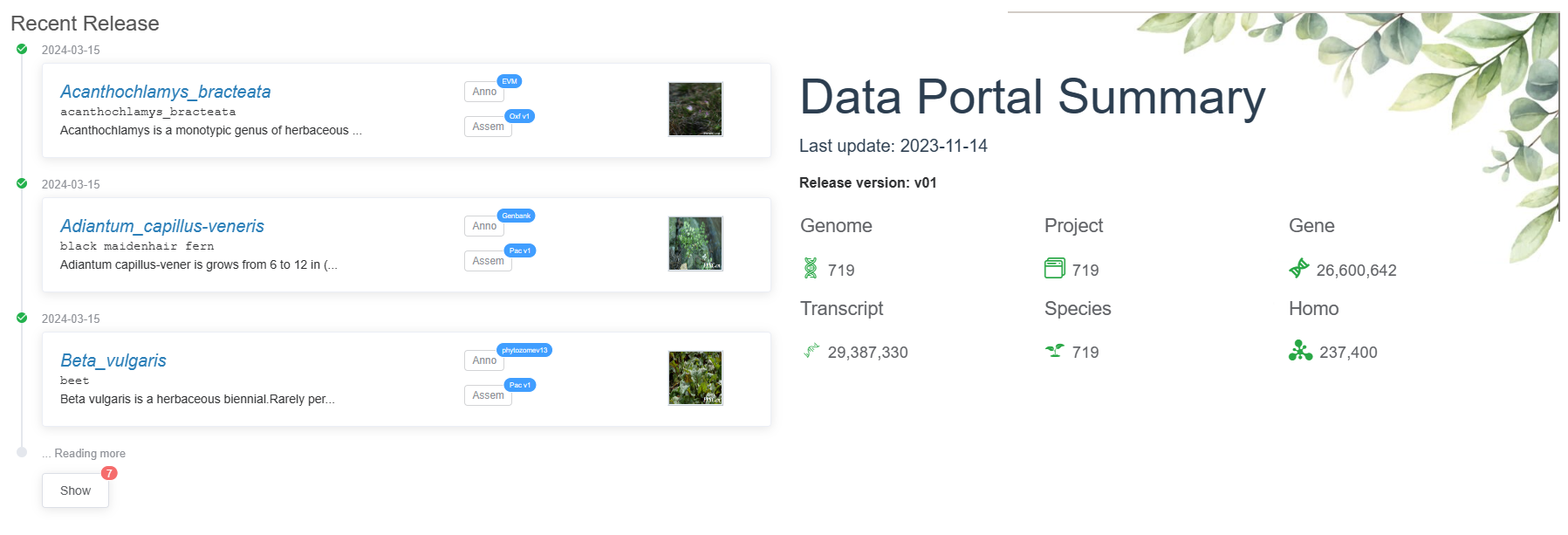
PGCP(Plant Genomes for Comparative Phylogenomics) is a central genomic datahub and a user-friendly analytic platform dedicated to facilitating scientific research in plant comparative studies. It serves as a central repository for genomic data, enabling researchers to access and analyze a vast array of plant genomic resources. Currently, PGdatabase hosts genomic resources from 719 diverse green plant species distributed across the major green plant species, including 9 rhodophytes, 1 glaucophytes, 1 prasinodermophytes, 39 chlorophytes, 7 charophytes, 131 bryophytes, 2 lycophytes, 5 ferns, 8 gymnosperms and 516 angiosperms. In total, there are 26,600,642 genes (29,387,330 transcripts). PGdatabase also provides predictions for orthogroups between proteins.
What does PGCP have?
PGCP mainly provides four modules:
Genome - Including image information for different species, detailed meta-information about the genome, ID information for Phytozome and NCBI taxonomy, transcription factor family information, download options for the .fa and .gff file formats of our species datasets and jump options for Blast and Browser, a tabular display of all the gene details of the species, and a table with the gene-by-gene detail pages and jump options for both Browsers (JBrowser and WashU Browser).
Download - Including genome files and genome annotation files for all PGCP species.
Tools - Including six essential tools: Blast, Browser, Functional Enrichment, ID Conversion, Synteny and Primer Designer.
Search - Including keyword search(PGCP ID and Source ID) and You have to identify the species.
Main features of PGCP
Comprehensive and Harmonized Data
PGCP is a comprehensive plant genome database that provides the largest collection of plant genomes and annotated datasets. The database currently includes 26,600,642 genes and 29,387,330 transcripts from 719 projects in 719 plant species and will be regularly updated with new datasets.
Interactive visualization
The dataset module can display the meta information of different species and show the basic information of all the genes of the species (including gene ID, location, positive and negative strand, TF information, GO annotation, functional annotation) through the table, and there are detail buttons for different genes to jump to the detailed information page to visualize the dataset easily.
Analysis tools
PGCP provides six different tools, including the basic blast function, Genome Browsers (Jbrowse2 and WashU Browser) for users to explore genomic data, Functional enrichment can help users to do Functional enrichment analysis, ID conversion tool can help users to do conversion between Plant Genome Catalog Gene ID and Source ID, Synteny tool can help users to view synteny for those genomes which have been assembled to chromsome level and Primer Designer tool can help users to design primers using Primer3.