3.1 Usage
See video - Quickly understand how to use the Search module through the following video tutorial.
Search - Users can find the search panel in the home page. The search panel provides an efficient way to help users quickly find the information they are looking for.
3.1.1 Basic usage
First select species - Users can select the species they are interested in from the drop-down list. The species list is based on the species that are available in the database. The default species is
Arabidopsis thaliana.Then input query in search box - Users can enter the
source gene IDorPlanT2T IDthey are looking for in the search box. The search box supports bothexact matchandfuzzy matchsearch. The search box will automatically prompt the user with the matching results as the user types in the search box.Finally click Search button

3.1.2 Exact match search
Input a full gene ID and click the search button for exact match search. If the gene ID is correct and exists in the database, the system will return the corresponding gene page diractly.
e.g. AT1G01010 is a gene ID of Arabidopsis thaliana Col-CEN.

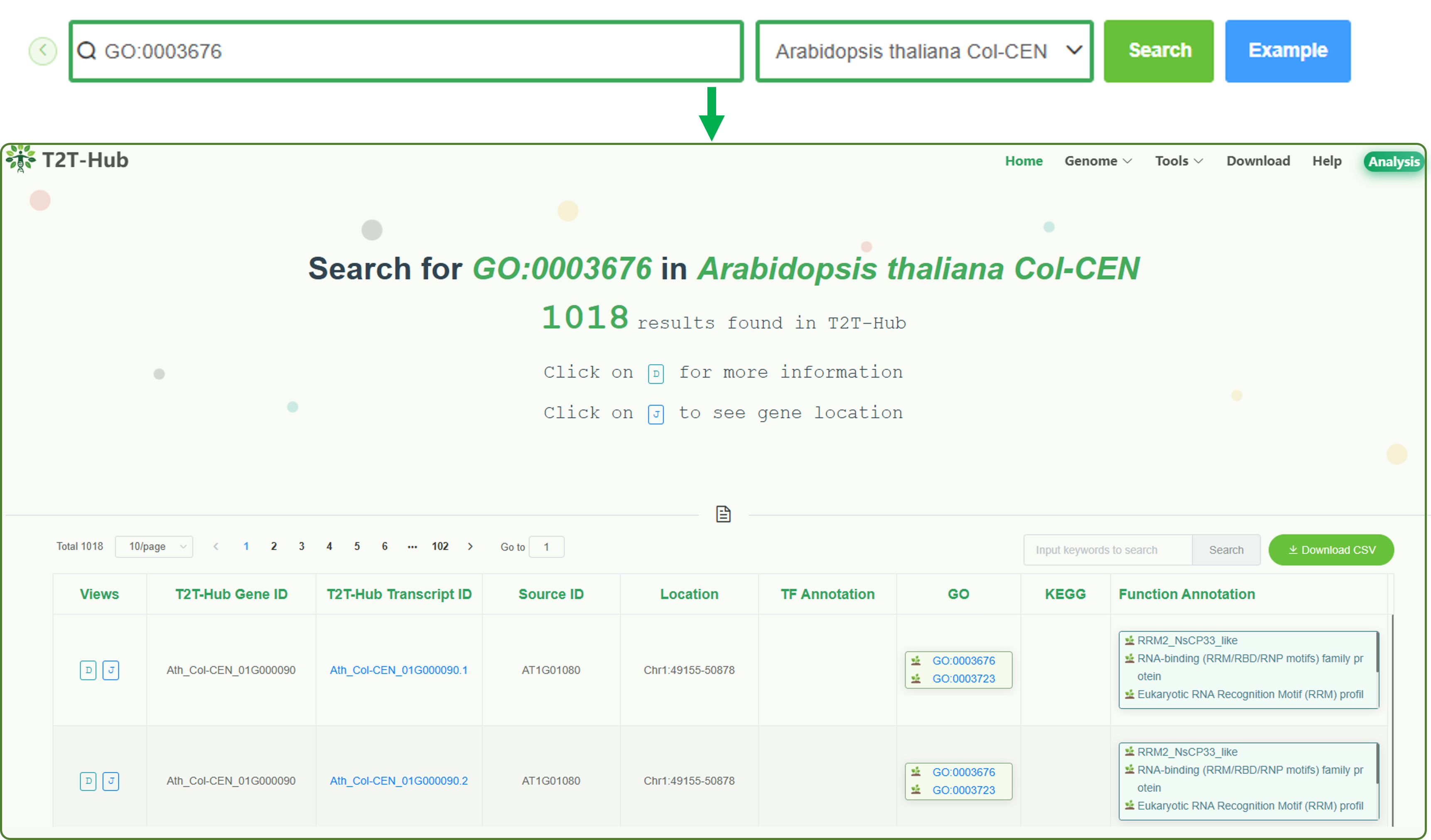
3.1.3 Fuzzy match
Input a part of gene ID or description (e.g. GO, KEGG, Function, etc.), then click the search button, the system will return multiple gene information that match the query, and the user can select the gene of interest from the search results to view the gene page.
e.g. GO:0003676 is a gene ontology term of Arabidopsis thaliana Col-CEN.
3.1.4 BLAST search
When users are not sure about the gene ID, they can use the BLAST search function to find the gene of interest. See how to use BLAST.

3.1.5 Genome search
PlanT2T provides a genome search function for users to search for the genome of interest. Then users can click the Genome name to view the genome page.